2. Pick particles with crYOLO#
This example covers crYOLO Tutorial 2, using particle picks from CryoSPARC as training data. The crYOLO GUI is not required for this tutorial.
To run this notebook, set up a conda environment as directed in the Running Example Notebooks instructions (be sure to run the optional crYOLO installation commands).
2.1. Prelude#
Connect to a cryoSPARC instance with cryosparc.tools.CryoSPARC and get the project handle. This project contains a workspace W3 with the following jobs:
Patch CTF with 20 motion-corrected and CTF-estimated micrographs
Exposures Sets tool that splits micrographs into 5 for training/validation and 15 for picking from the trained model
Manual Picker that picks 5 training micrographs to completion
Note
Saving crYOLO outputs to the CryoSPARC project directory requires file-system access to the project directory.
from cryosparc.tools import CryoSPARC
cs = CryoSPARC(host="cryoem0.sbi", base_port=61000)
assert cs.test_connection()
project = cs.find_project("P251")
Connection succeeded to CryoSPARC API at http://cryoem0.sbi:61002
Programatically create and build a new external job. This job will contain the results of both training and picking for the remaining micrographs.
Use the job.connect function to connect the training picks, training micrographs and remaining micrographs as inputs. This function has the following signature:
connect(
target_input: str,
source_job_uid: str,
source_output: str,
slots: list[SlotSpec] = [],
title: str = "",
desc: str = "",
)
Where,
target_inputis the name of the input to create on the new job, e.g.,"train_particles"source_job_uidis the unique ID of the parent job to connect for this input, e.g.,"J42"source_outputis the name of the output on the parent jobslots(optional) is a list of required results in the parent output.
Specify micrograph_blob as slots for the micrographs to retrieve the motion-corrected micrograph path. Other micrograph slots will be connected as passthroughs. Specify the location slot to retrieve the \((x, y)\) coordinates for each particle in their original micrograph.
Create an output slots for the resulting picks. It must inlude location and
pick_stats slots
job = project.create_external_job("W3", title="crYOLO Picks")
job.connect("train_micrographs", "J18", "split_0", slots=["micrograph_blob"])
job.connect("train_particles", "J19", "particles_selected", slots=["location"])
job.connect("all_micrographs", "J18", "split_0", slots=["micrograph_blob"])
job.connect("all_micrographs", "J18", "remainder", slots=["micrograph_blob"])
job.add_output("particle", "predicted_particles", slots=["location", "pick_stats"])
'predicted_particles'
Read the Jobs Guide for instructions on inspecting job outputs to determine their names and available slots for use in this connection step.
Start the job to write to the outputs and job log. This puts the job in “Waiting” status.
job.start()
2.2. Data Preparation#
Use the job directory as crYOLO’s working directory. Create all the necessary subfolders there.
job.mkdir("full_data")
job.mkdir("train_image")
job.mkdir("train_annot")
Load the input micrographs and link them into the full_data and train_image directories. This results in the following directory structure:
/path/to/project/JX/
├── full_data
│ ├── mic01.mrc -> /path/to/project/JY/motioncorrected/mic01.mrc
│ ├── mic02.mrc -> /path/to/project/JY/motioncorrected/mic02.mrc
│ ├── ...
│ └── mic20.mrc -> /path/to/project/JY/motioncorrected/mic20.mrc
└── train_image
├── mic01.mrc -> /path/to/project/JY/motioncorrected/mic01.mrc
├── mic02.mrc -> /path/to/project/JY/motioncorrected/mic02.mrc
├── mic03.mrc -> /path/to/project/JY/motioncorrected/mic03.mrc
├── mic04.mrc -> /path/to/project/JY/motioncorrected/mic04.mrc
└── mic05.mrc -> /path/to/project/JY/motioncorrected/mic05.mrc
all_micrographs = job.load_input("all_micrographs", ["micrograph_blob"])
train_micrographs = job.load_input("train_micrographs", ["micrograph_blob"])
for mic in all_micrographs.rows():
source = mic["micrograph_blob/path"]
target = job.uid + "/full_data/"
project.symlink(source, target)
for mic in train_micrographs.rows():
source = mic["micrograph_blob/path"]
target = job.uid + "/train_image/"
project.symlink(source, target)
crYOLO requires the particle locations for each micrograph in STAR format with the following directory stucture:
/path/to/project/JX/
└── train_annot
└── STAR
├── mic01.star
├── mic02.star
├── mic03.star
├── mic04.star
└── mic05.star
Load the training particle locations. Split them up my micrograph path. Compute the pixel locations and save them to a star file in this format.
from io import StringIO
import numpy as np
from cryosparc import star
job.mkdir("train_annot/STAR")
train_particles = job.load_input("train_particles", ["location"])
for micrograph_path, particles in train_particles.split_by("location/micrograph_path").items():
micrograph_name = micrograph_path.split("/")[-1]
star_file_name = micrograph_name.rsplit(".", 1)[0] + ".star"
mic_w = particles["location/micrograph_shape"][:, 1]
mic_h = particles["location/micrograph_shape"][:, 0]
center_x = particles["location/center_x_frac"]
center_y = particles["location/center_y_frac"]
location_x = center_x * mic_w
location_y = center_y * mic_h
outfile = StringIO()
star.write(
outfile,
np.rec.array([location_x, location_y], names=["rlnCoordinateX", "rlnCoordinateY"]),
)
outfile.seek(0)
job.upload("train_annot/STAR/" + star_file_name, outfile)
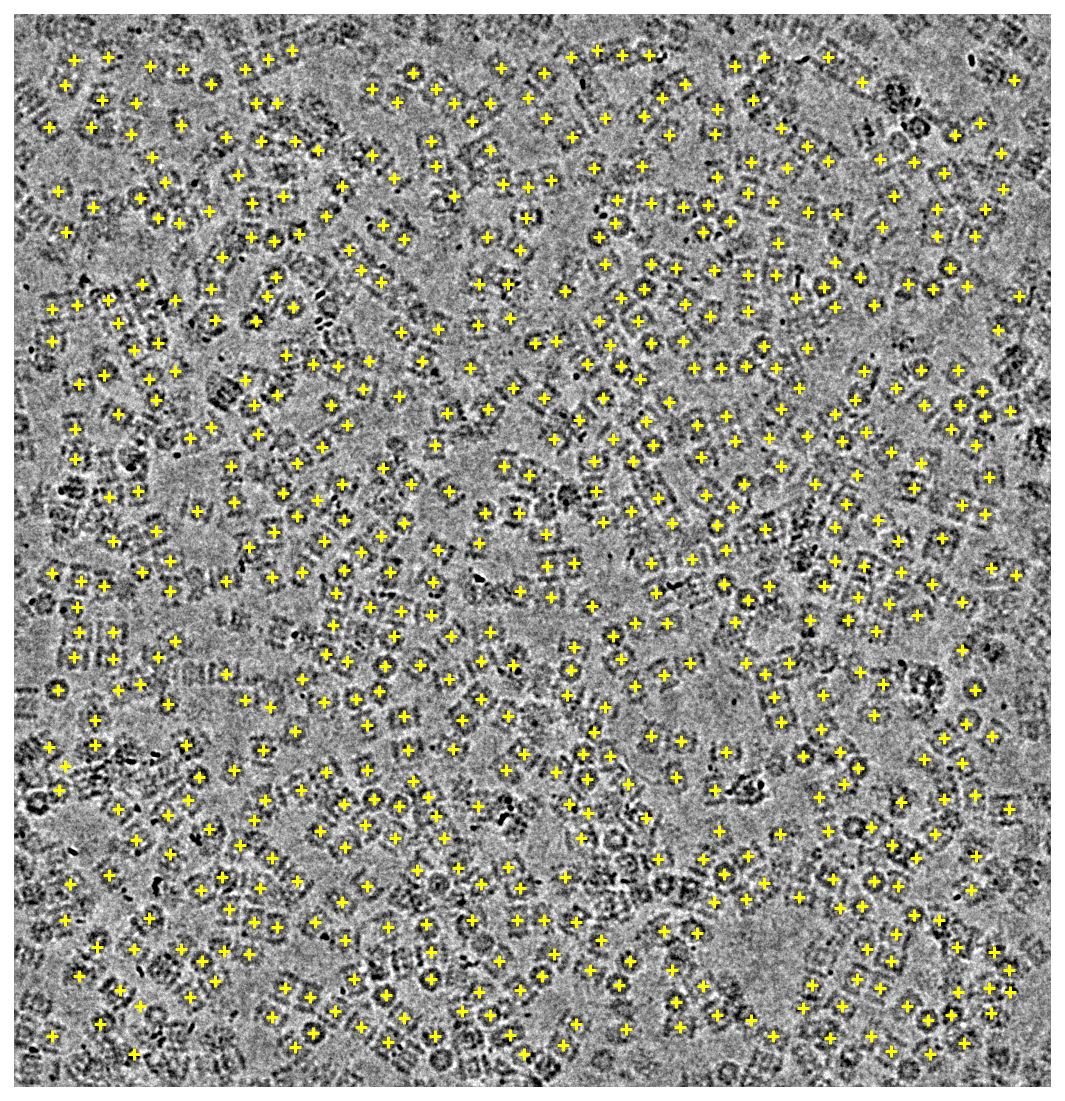
Preview of the particle locations used on the last training micrograph:
%matplotlib inline
import matplotlib.pyplot as plt
from cryosparc.tools import downsample, lowpass2
header, mic = project.download_mrc(micrograph_path)
binned = downsample(mic, factor=3)
lowpassed = lowpass2(binned, psize_A=0.6575, cutoff_resolution_A=20, order=0.7)
height, width = lowpassed.shape
vmin = np.percentile(lowpassed, 1)
vmax = np.percentile(lowpassed, 99)
fig, ax = plt.subplots(figsize=(7.5, 8), dpi=144)
ax.axis("off")
ax.imshow(lowpassed, cmap="gray", vmin=vmin, vmax=vmax, origin="lower")
ax.scatter(center_x * width, center_y * height, c="yellow", marker="+")
fig.tight_layout()
2.3. Configuration#
cryosparc-tools provides a job.subprocess function to run arbitrary processes, including cryolo_*.py scripts installed in the active conda environment.
Use job.subprocess to generate a crYOLO configuration file with the cryolo_gui.py config command. Specify a box size of 130 for this dataset.
Note
When connecting to a remote CryoSPARC instance, note that job.subprocess processes will run on the local machine, not remotely.
job.subprocess(
(
"cryolo_gui.py config config_cryolo.json 130 --train_image_folder train_image --train_annot_folder train_annot"
).split(" "),
cwd=job.dir,
)
#####################################################
Important debugging information.
In case of any problems, please provide this information.
#####################################################
/u/nfrasser/micromamba/envs/cryolo/bin/cryolo_gui.py config config_cryolo.json 130
--train_image_folder train_image
--train_annot_folder train_annot
#####################################################
Wrote config to config_cryolo.json
job.subprocess also forwards the output to the job stream log. Arguments such as cwd are similar to Python’s subprocess.Popen().
This results in a config_cryolo.json configuration file in the external job directory.
2.4. Training#
Run training on GPU 0 with 5 warmup-epochs and an early stop of 15.
The output of this command is quite long, so set mute=True to hide it (it will still appear in the job’s Event log).
Use the checkpoint_line_pattern flag new training epoch lines as the beginning of a checkpoint in the Event log.
job.subprocess(
"cryolo_train.py -c config_cryolo.json -w 5 -g 0 -e 15".split(" "),
cwd=job.dir,
mute=True,
checkpoint=True,
checkpoint_line_pattern=r"Epoch \d+/\d+", # e.g., "Epoch 42/200"
)
Open the External job’s “Events” tab from CryoSPARC’s web interface to view crYOLO’s output. When the process completes, crYOLO creates a cryolo_model.h5 trained model file in the job directory.
2.5. Picking#
Use the trained model to predict particle locations for the full dataset. Create a boxfiles directory to store the output.
job.mkdir("boxfiles")
job.subprocess(
"cryolo_predict.py -c config_cryolo.json -w cryolo_model.h5 -i full_data -g 0 -o boxfiles -t 0.3".split(" "),
cwd=job.dir,
mute=True,
checkpoint=True,
)
For each micrograph in the full dataset, load the corresponding output particles star file, initialize a new empty particles dataset and fill in the predicted locations and other relevant location metadata.
Also fill in a dummy NCC score so that the results may be inspected with an Inspect Picks job.
output_star_folder = "STAR"
all_predicted = []
for mic in all_micrographs.rows():
micrograph_path = mic["micrograph_blob/path"]
micrograph_name = micrograph_path.split("/")[-1]
height, width = mic["micrograph_blob/shape"]
starfile_name = micrograph_name.rsplit(".", 1)[0] + ".star"
starfile_path = "boxfiles/STAR/" + starfile_name
locations = star.read(job.dir / starfile_path)[""]
center_x = locations["rlnCoordinateX"] / width
center_y = locations["rlnCoordinateY"] / height
predicted = job.alloc_output("predicted_particles", len(locations))
predicted["location/micrograph_uid"] = mic["uid"]
predicted["location/micrograph_path"] = mic["micrograph_blob/path"]
predicted["location/micrograph_shape"] = mic["micrograph_blob/shape"]
predicted["location/micrograph_psize_A"] = mic["micrograph_blob/psize_A"]
predicted["location/center_x_frac"] = center_x
predicted["location/center_y_frac"] = center_y
predicted["pick_stats/ncc_score"] = 0.5
all_predicted.append(predicted)
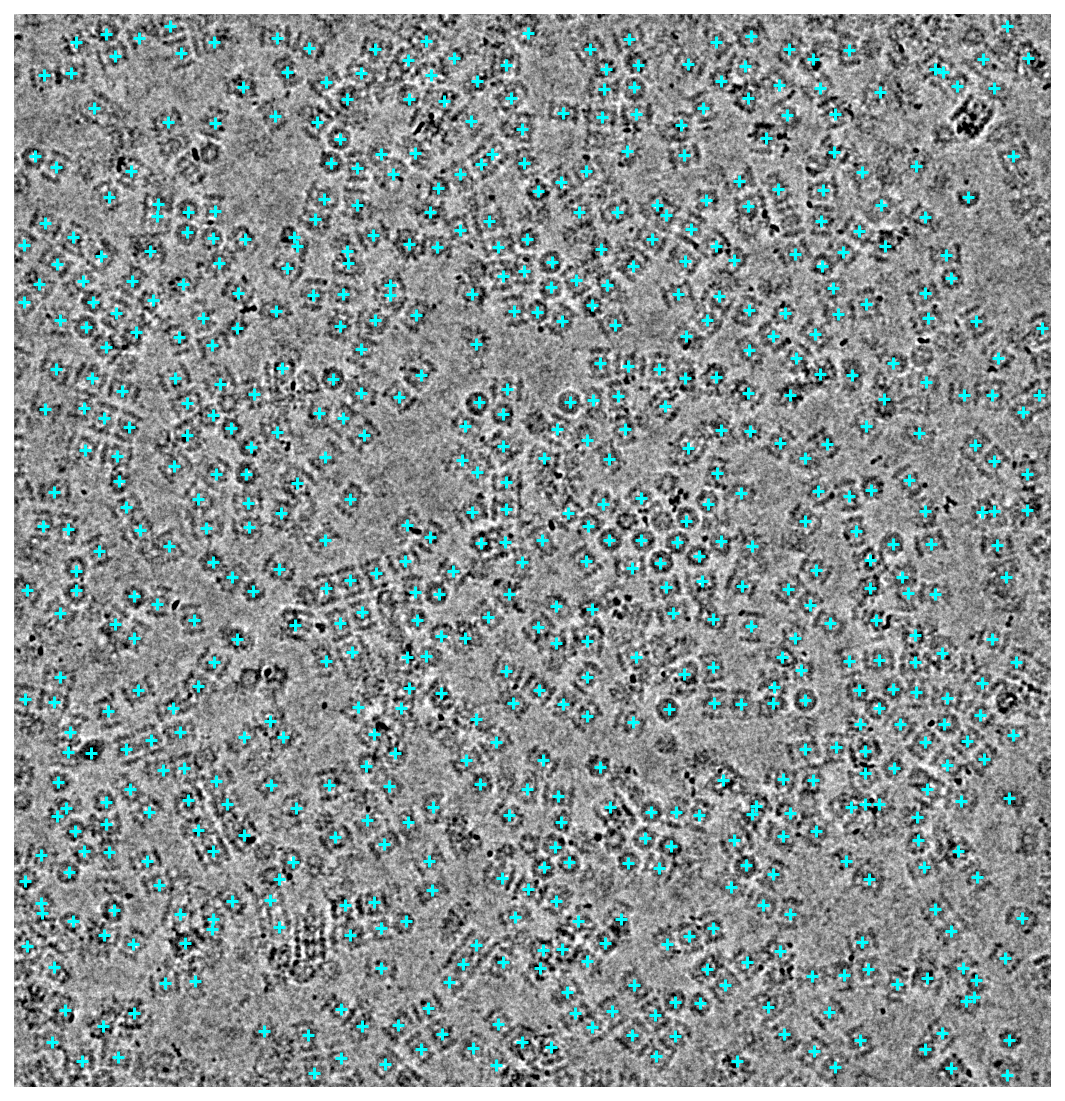
Output the most recent predicted particle locations to verify that crYOLO ran successfully.
header, mic = project.download_mrc(micrograph_path)
binned = downsample(mic, factor=3)
lowpassed = lowpass2(binned, psize_A=0.6575, cutoff_resolution_A=20, order=0.7)
height, width = lowpassed.shape
vmin = np.percentile(lowpassed, 1)
vmax = np.percentile(lowpassed, 99)
fig, ax = plt.subplots(figsize=(7.5, 8), dpi=144)
ax.axis("off")
ax.imshow(lowpassed, cmap="gray", vmin=vmin, vmax=vmax, origin="lower")
ax.scatter(center_x * width, center_y * height, c="cyan", marker="+")
fig.tight_layout()
Append all the predicted particles into a single dataset. Save this to the job and mark as completed.
from cryosparc.dataset import Dataset
job.save_output("predicted_particles", Dataset.append(*all_predicted))
job.stop()
