3. 3D Flex: Custom Latent Trajectory#
Note
Read the tutorial to learn more about 3D Flexible Refinement.
This example covers the following:
Load particle latent coordinates from a 3D Flex Training job
Plot the latent coordinates
Interactively draw a trajectory through the latent space
Output the trajectory as a new output in CryoSPARC
The resulting trajectory may be used as input to the 3D Flex Generator job to generate a volume series along the trajectory. In this way, you can visualize specific regions or pathways through the latent conformational distribution of the particle.
import json
from pathlib import Path
import numpy as n
from cryosparc.tools import CryoSPARC
with open(Path("~", "instance-info.json").expanduser(), "r") as f:
credentials = json.load(f)
cs = CryoSPARC(**credentials)
assert cs.test_connection()
Connection succeeded to CryoSPARC command_core at http://cryoem0.sbi:40002
Connection succeeded to CryoSPARC command_vis at http://cryoem0.sbi:40003
Connection succeeded to CryoSPARC command_rtp at http://cryoem0.sbi:40005
Load the particles dataset from the 3D Flex Training job.
import pandas as pd
project = cs.find_project("P312")
particles = project.find_job("J243").load_output("particles")
# only display the first ten rows, since creating an entire dataframe is slow
pd.DataFrame(particles.rows()[:10])
| alignments2D/alpha | alignments2D/alpha_min | alignments2D/class | alignments2D/class_ess | alignments2D/class_posterior | alignments2D/cross_cor | alignments2D/error | alignments2D/error_min | alignments2D/image_pow | alignments2D/pose | ... | pick_stats/power | pick_stats/template_idx | sym_expand/helix_num_rises | sym_expand/helix_rise_A | sym_expand/helix_twist_rad | sym_expand/idx | sym_expand/is_helix | sym_expand/src_uid | sym_expand/symmetry | uid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 1.067518 | 8 | 1.000261 | 0.999869 | 76.271484 | 9163.180664 | 0.0 | 9203.728516 | 4.744446 | ... | 781.205933 | 3 | 0 | 0.0 | 0.0 | 0 | 0 | 6947839038024507105 | C2 | 6947839038024507105 |
| 1 | 1.0 | 1.067518 | 8 | 1.000261 | 0.999869 | 76.271484 | 9163.180664 | 0.0 | 9203.728516 | 4.744446 | ... | 781.205933 | 3 | 0 | 0.0 | 0.0 | 1 | 0 | 6947839038024507105 | C2 | 13932325671802011693 |
| 2 | 1.0 | 0.868515 | 2 | 1.006742 | 0.996642 | 62.421875 | 8964.066406 | 0.0 | 8990.552734 | 3.205707 | ... | 596.804443 | 3 | 0 | 0.0 | 0.0 | 0 | 0 | 18174470766076440683 | C2 | 18174470766076440683 |
| 3 | 1.0 | 0.868515 | 2 | 1.006742 | 0.996642 | 62.421875 | 8964.066406 | 0.0 | 8990.552734 | 3.205707 | ... | 596.804443 | 3 | 0 | 0.0 | 0.0 | 1 | 0 | 18174470766076440683 | C2 | 16990665930294442579 |
| 4 | 1.0 | 1.158005 | 0 | 1.656703 | 0.749064 | 47.288086 | 8603.187500 | 0.0 | 8630.057617 | 5.385587 | ... | 758.841248 | 3 | 0 | 0.0 | 0.0 | 0 | 0 | 10868047345897440647 | C2 | 10868047345897440647 |
| 5 | 1.0 | 1.158005 | 0 | 1.656703 | 0.749064 | 47.288086 | 8603.187500 | 0.0 | 8630.057617 | 5.385587 | ... | 758.841248 | 3 | 0 | 0.0 | 0.0 | 1 | 0 | 10868047345897440647 | C2 | 13431911868430011898 |
| 6 | 1.0 | 0.992612 | 6 | 1.032084 | 0.984298 | 39.340820 | 8956.469727 | 0.0 | 8975.994141 | 0.000000 | ... | 601.065002 | 3 | 0 | 0.0 | 0.0 | 0 | 0 | 11087656592019060700 | C2 | 11087656592019060700 |
| 7 | 1.0 | 0.992612 | 6 | 1.032084 | 0.984298 | 39.340820 | 8956.469727 | 0.0 | 8975.994141 | 0.000000 | ... | 601.065002 | 3 | 0 | 0.0 | 0.0 | 1 | 0 | 11087656592019060700 | C2 | 3521966667339501106 |
| 8 | 1.0 | 0.992270 | 8 | 1.000796 | 0.999602 | 71.224609 | 8801.692383 | 0.0 | 8837.027344 | 0.128228 | ... | 669.910767 | 3 | 0 | 0.0 | 0.0 | 0 | 0 | 10759805808414285613 | C2 | 10759805808414285613 |
| 9 | 1.0 | 0.992270 | 8 | 1.000796 | 0.999602 | 71.224609 | 8801.692383 | 0.0 | 8837.027344 | 0.128228 | ... | 669.910767 | 3 | 0 | 0.0 | 0.0 | 1 | 0 | 10759805808414285613 | C2 | 2825550586744122481 |
10 rows × 110 columns
In this notebook, we create a plot of two of the dimensions and click to generate the trajectory through latent space. This approach necessarily allows only the creation of a trajectory through two of the latent coordinates. If you need to change three or more coordinates simultaneously, you will have to manually create a list of the coordinates you want to use.
selected_dimensions = [0, 2]
Next we create an interative plot that responds to on-click events.
Once the plot is drawn, click repeatedly on the plot along a trajectory you wish to sample in the latent space. The points along this trajectory will form the output of the notebook and be used in 3D Flex Generator.
Note
Using interactive plots in Jupyter Notebooks can present some challenges.
Connecting to a remote VS Code remote notebook requires the ipympl package
and the %matplotlib widget “magic” line.
See the matplotlib documentation
for more detail.
%matplotlib widget
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(5, 5))
def do_plot():
plt.plot(
particles[f"components_mode_{selected_dimensions[0]}/value"][::10],
particles[f"components_mode_{selected_dimensions[1]}/value"][::10],
".",
alpha=0.5,
color="gray",
)
plt.grid()
do_plot()
pts = []
def onclick(event):
fig.clf()
do_plot()
pts.append([event.xdata, event.ydata])
apts = n.array(pts)
plt.plot(apts[0, 0], apts[0, 1], "xk")
plt.plot(apts[:, 0], apts[:, 1], ".-r")
cid = plt.gcf().canvas.mpl_connect("button_press_event", onclick)
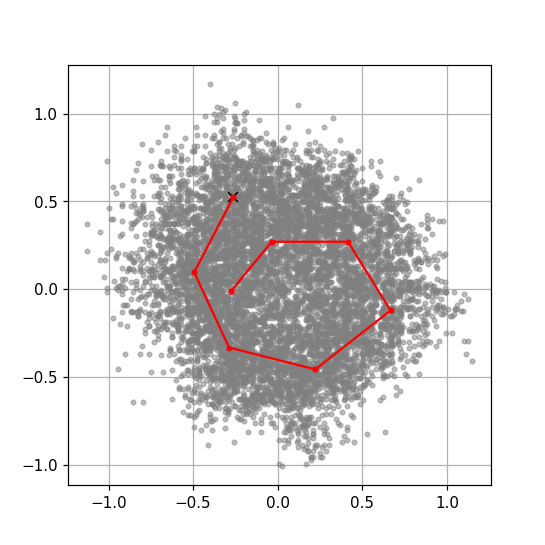
Print out the selected trajectory points. Each column corresponds to the position along that selected coordinate, i.e., in this example column 1 is coordinate 0 and column 2 is coordinate 2.
latent_pts = n.array(pts)
for pt in latent_pts:
print(f"{pt[0]:5.2f}, {pt[1]:5.2f}")
-0.35, 0.67
-0.78, 0.09
-0.50, -0.67
0.77, -0.86
1.06, -0.18
0.95, 0.56
0.04, 0.43
-0.27, -0.11
Set up an external job to save the custom latent components. Connect to the train job to ensure output fields get passed through.
# each component has a components_mode_n/component and components_mode_n/value field,
# so we need to divide the components_mode fields by two to get the total number of components
num_components = int(len([x for x in particles.fields() if "components_mode" in x]) / 2)
slot_spec = [{"dtype": "components", "name": f"components_mode_{k}"} for k in range(num_components)]
job = project.create_external_job("W5", "Custom Latents")
job.connect("particles", "J243", "particles", slots=slot_spec)
True
Add and allocate an output for the job to store the custom latent components.
latents_dset = job.add_output(
type="particle",
name="latents",
slots=slot_spec,
title="Latents",
alloc=len(latent_pts),
)
Save the points into the allocated dataset.
for k in range(num_components):
latents_dset[f"components_mode_{k}/component"] = k
try:
latents_dset[f"components_mode_{k}/value"] = latent_pts[:, selected_dimensions.index(k)]
except ValueError:
# if a coordinate is not in our selected_dimensions, set the trajectory to zero in that coordinate
latents_dset[f"components_mode_{k}/value"] = 0
Save the output.
with job.run():
job.save_output("latents", latents_dset)
